Introduction
Cryobank of pathogenic bacterial strains isolated from periprosthetic and surgical infections
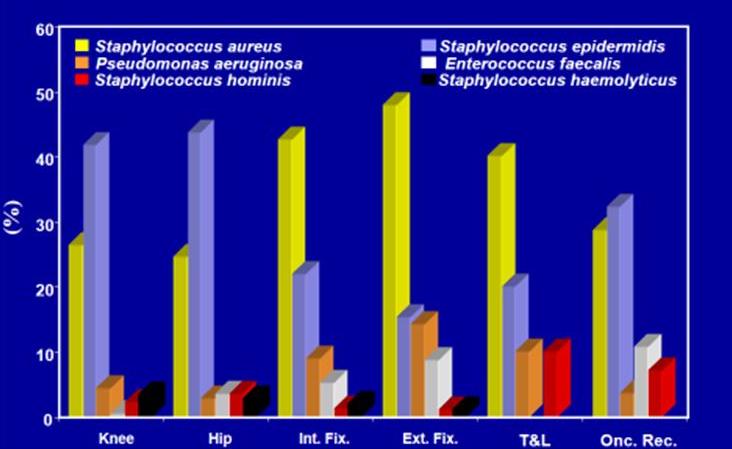
A strain library has been created where the bacterial clinical isolates, responsible of periprosthetic and surgical infections diagnosed and treated at the Rizzoli Orthopaedic Institute, are stored.
The bacteriologic samples are processed following a protocol optimised to ensure an accurate identification, labelling, control, viability and storage of the microbial strains deposited.

1. Size of the strain library
Over 2.700 clinical isolates, obtained from patients undergoing revision of surgical infections and treatment of infected prostheses at the Rizzoli Orthopaedic Institute, have currently been collected. Each single strain is stored at -80°C in cryoresistant vials, suspended in a medium consisting of 90% nutrient broth and 10% glycerol. The culture of each single strain is aliquoted and stored in multiple vials, part of which are reserved to back-ups. The samples are progressively numbered and labelled by a code in order to ensure the respect of the laws on privacy.
2. Identification
The identification, conducted by conventional biochemical galleries such as API, is further verified by automated ribotyping using the RiboPrinter® (DuPont) of the Research Unit on Implant Infections. The identification is performed down to a species, subspecies and ribotype level.
3. Antibioticresistance
The sensitivity to antibiotics is assayed by the agar disk diffusion method (Kirby-Bauer) according to the guide lines of the National Committee for
Clinical Laboratory Standards Institute (CLSI). Disks of 16 different antibiotic drugs, at the concentrations recommended by NCCLS, are utilised. The diameters of the areas of growth inhibition are measured in mm by a dedicated image-analysis system (Biovideobact, Biokit), which automatically performs the measurement of the halos and, if necessary, allows to determine the MIC for each single antibiotic.
4. Phenotypic characterisation of the biofilm production
The strains are assessed for their capability to form biofilm. In the case of staphylococci the evaluation was performed by the Congo red-agar (CRA) test, following an optimisation of the original procedure setup at the Laboratory on Implant Infections (Arciola et al., Biomaterials, 2002;23:4233-9). Currently, new methods for the quantification and characterization of the different biofilm macromolecular components have been developed at our Laboratory.
5. Genotypic characterisation
The strains are analysed by molecular biology techniques (PCR with specific primers and DNA-hybridisation) for the presence of the genes mediating the production of biofilm polysaccharide (genes of the locus ica), the genes of the adhesins that bind to host matrix proteins (also named MSCRAMMs) and the genes of toxins.
6. Ribotyping
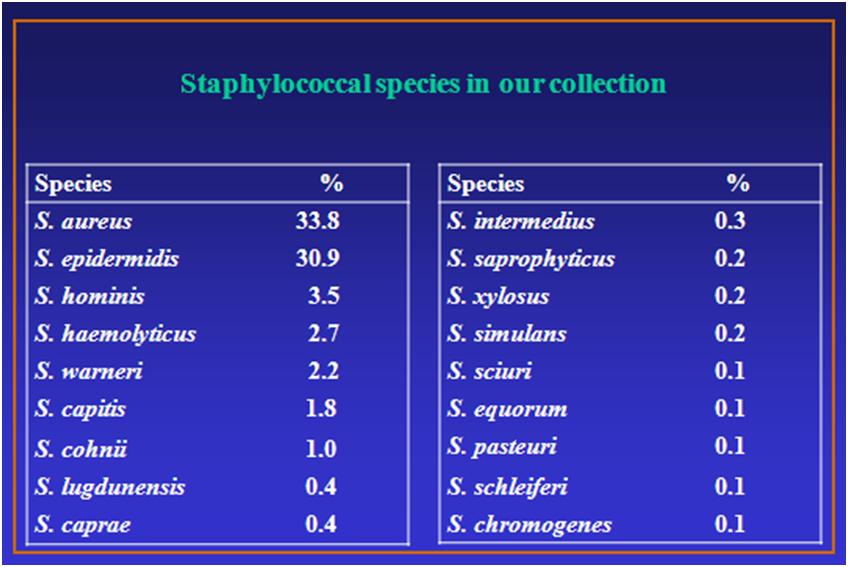
Bacterial species isolated and stored in the strain library
The identification by RiboPrinter® is performed especially on rare species, usually opportunistic, or pathogens of animal origin (S. caprae, S. equorum and others), comparing the ribotype patterns with those of the DuPont archives and defining the ribogroup. Particular attention is paid to the collection of coagulase-negative staphylococci that are currently emerging as 'new pathogens'.
In the case of the main etiologic agents of infections associated with orthopedic implants, the ribo-typing is currently utilized to identify the epidemic clones.