Introduction
The identification of the bacterial isolates by the use of conventional phenotypic techniques such as the biochemical galleries, widely utilised by common diagnostic microbiology laboratories, is often inadequate for studies of molecular pathogenesis and epidemiology, where a sure identification of the bacterium not only at the level of species, but often of subspecies if not even of clone is required.
Thus, it is of fundamental importance in the studies performed at the Laboratory on Implant Infections to take advantage of techniques of greater reliability and based on genotypic typing of the isolates. In this regard, ribotyping represents a recent and very accredited technique of genotypic typing, based on the analysis of polymorphisms of restriction fragments of the genes encoding for the ribosomal RNAs (rRNA 23S, 16S e 5S).
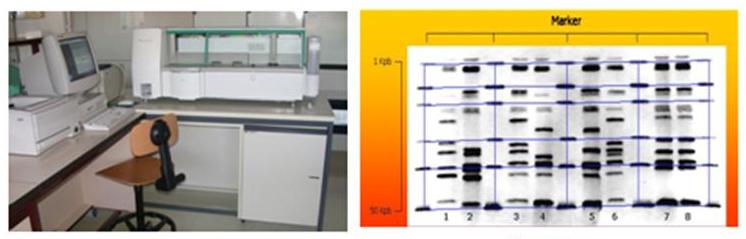
The RiboPrinter® Microbial Characterization System (DuPont-Qualicon) of the Research Unit on Implant Infections allows to perform the genotypic typing of the isolates through a process of automated ribotyping, also referred to as 'riboprinting'. It represents a further evolution with respect to the manual techniques of ribotyping, long known and characterised by high laboriousness and, sometimes, limited reproducibility.

In practice, the RiboPrinter®, requiring just a minimal initial intervention by the operator, performs all the following actions: bacterial lysis, DNA extraction, digestion by restriction enzymes, electrophoretic separation of restriction fragments, transfer on membrane, membrane processing and hybridisation with a chemiluminescent labelled probe, capture and digitalization of the chemiluminescent signal, elaboration of the image in a ribotyping profile, comparison of the RiboPrint patterns to DuPont identification library, taxonomic identification of the isolate and analysis of the similarity with respect to reference strain profiles present in the DuPont archive, attribution to specific ribogroups based on the similarity to the samples previously analysed at the Laboratory on Implant Infections.
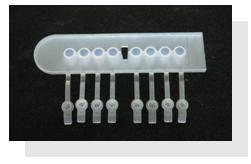
The sample holder includes 8 different tracks where to insert 8 different isolates, which will be processed simultaneously. Every isolates from the same sample holder is considered as belonging to the same analysis batch.